With the three-year SARS‑CoV‑2 coronavirus crisis barely in the rear-view mirror, experts agree it’s only a matter of time before we are hit by another global pandemic.
To be ready for that inevitable arrival, an international crowdsourced campaign to discover an anti-Covid-19 drug has created a blueprint for the accelerated, patent-free development of drugs to treat future viral threats.
The results of the COVID Moonshot campaign, published November 9 in Science, include more than 18,000 compound designs, more than 2,400 synthesized compounds, more than 500 3D structures obtained by X-ray crystallography and more than 10,000 measurements from biochemical assays.
Pharmaceutical companies can use this “open science” platform to develop inexpensive generic drugs. The international nonprofit organization Drugs for Neglected Diseases Initiative, which aids poor and vulnerable communities, has adopted COVID Moonshot into its drug-development portfolio.
Co-led by Prof. Nir London of the Weizmann Institute of Science’s Chemical and Structural Biology Department with researchers from the University of Oxford, Memorial Sloan Kettering Cancer Center and the US-based biotech company PostEra, the campaign has already helped create a brand-name medication.
The anti-COVID-19 drug Xocova (ensitrelvir), which received emergency approval in Japan in 2022, was developed in part on the basis of crystallographic data from the consortium.

The obvious thing to do
While working to discover effective antiviral drugs during the Covid-19 pandemic, London and his international colleagues decided to allow open access to their findings, posting them online in real time for the benefit of the entire scientific community.
“At the time of a global crisis, it seemed like the obvious thing to do,” London said.
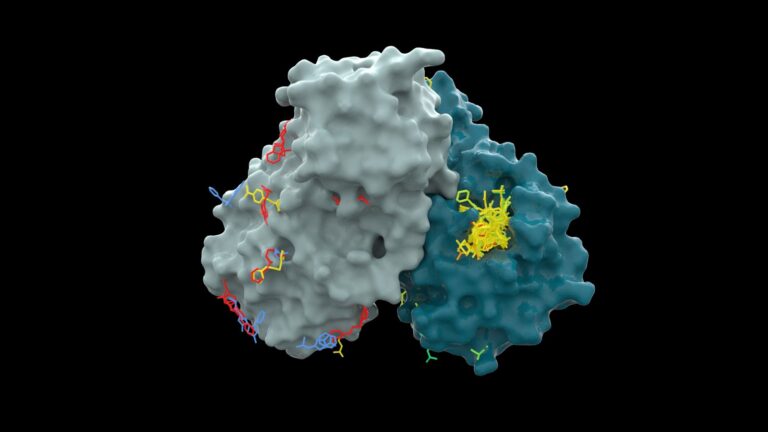
He was collaborating at the time with researchers from the University of Oxford and the UK’s national synchrotron Diamond Light Source, who had mapped the 3D structure of a key COVID-19 protein and were seeking a molecule that would block this enzyme.
They then teamed up with PostEra, using its machine learning tools to identify the most promising of these compounds and to look for new ones.
On Twitter (now called X), London invited medicinal chemists and computer-aided drug design experts to suggest how the original compounds could be improved. Within the first week alone, the team received more than 2,000 submissions.
“Many scientists were in lockdown, and they were only too happy to enlist in a worthy cause,” London said.
“That was when I realized that ‘science for the future of humanity’ is not only a Weizmann Institute tagline – it’s become a reality.”
Global collaboration
A volunteer team of medicinal chemists, eventually led by Ed Griffen of the UK-based biotech company MedChemica, triaged the compounds and continued to design new ones.
A team from Memorial Sloan Kettering Cancer Center led the computational evaluation of the submissions, and a company in Ukraine, Enamine, synthesized the compounds predicted to be most effective against Covid-19.
Packages with these molecules started arriving at Weizmann on a weekly basis. London tested them at the Nancy and Stephen Grand Israel National Center for Personalized Medicine on the Weizmann campus.
The results were posted online and numerous researchers suggested further improvements; the improved molecules were again synthesized and tested.
The optimization process progressed in this manner for more than 50 iterations.
The initial collaboration ultimately grew into the much larger COVID Moonshot Consortium, which received $10 million in funding from the Wellcome Trust. The researchers donated their time, ideas and resources, and all their data was published immediately available without intellectual property restrictions.
The group’s leaders have founded an antiviral drug discovery center, which recently received initial funding of $68 million from the United States National Institutes of Health.
The center will identify compounds that can be developed into drugs for three large families of viruses – coronaviruses, picornaviruses and flaviviruses – that threaten to cause future pandemics.
“The COVID Moonshot has demonstrated that open science offers an effective alternative model for drug discovery,” London said.
“By building on the strength of numerous contributors and removing intellectual property barriers and red tape, it can tremendously speed up the development of drugs that are urgently needed to avert global threats.”